Picograms of Double Stranded RNA a Headache for Jab Profiteers
Australia's Therapeutic Goods Adminstration has a special interest in dsRNA but mainstream media won't tell you about it.
FOI documents obtained from Australia's Therapeutic Goods Administration (TGA) show some of the Pfizer Lots where they measured dsRNA that looks like this.1
Previously I wrote that Katalin Karikó of BioNTech and Pfizer (and Nobel Prize fame) studied how the Human body responds to foreign dsRNA and mRNA and they deliberately targeted Toll-Like Receptor 3 (TLR3) in Dendritic cells in 2004.2
I mentioned Moderna jabs causing Agranulocytosis, the collapse of white blood cells often associated with Leukemia, and this is known to proceed via dsRNA triggering TLR3. Please see reference 10 in my Leukemia article.3
Legionnaires Disease also proceeds via dsRNA.4
A number of my friends are interested, including Maria Gutschi5, seen here replying to David Speicher. dsRNA is definitely a worry but less toxic than Endotoxin.6
Maria led me to a very useful review of the dsRNA problem.7
David Cowley has invested a lot of effort explaining the hazards of dsRNA.8
Our mutual friend Kevin McKernan says9, comparing Moderna with Pfizer:
That’s just a DNA perspective. I suspect they have move dsRNA which is harder to measure but predicted by RNAfold of their mRNA. Their codon optimization is more GC rich than Pfizer and has more secondary structure. Also a higher 100ug dose.
After posting, I discovered our friend Patent Sun10 wrote a detailed Substack in May 2024 specifically about dsRNA and what he discovered by reading patents.11
One of the major worries about dsRNA concerns the enzyme RNA polymerase III which plays an essential role in detecting foreign DNA (non-self nucleic acids) which transcribes the ssDNA into double-stranded RNA (dsRNA) containing a 5′-triphosphate moiety that causes the devastating toxic consequences.12
Zika virus uses dsRNA to replicate
Understanding dsRNA is helped by studies of how viruses other than Covid19 invade and multiply, as found by a multinational team of researchers.13
Here is their caption:
Fig. 2 Schematic overview for ZIKV replication cycle. E protein interacts with host-cell receptors and the virus enters cells by receptor-mediated endocytosis. Several ZIKV receptors were identified including AXL, Tyro3, DC-SIGN, and TIM-1. Into the endosome occurs the fusion of viral particles with the endosomal membrane by and acidic-pH-triggered mechanism followed by virus disassembly and release of viral RNA into the cytoplasm. The (+)ssRNA is translated into a polyprotein further processed and cleaved into all structural (C, prM, and E) and non-structural proteins (NS1 to NS5). The replication process occurs at the surface of the endoplasmic reticulum (ER). A dsRNA genome is synthesized from the genomic (+)ssRNA. Transcription and replication of the dsRNA genome provide viral mRNAs and new (+)ssRNA. Virus assembly occurs at the ER and leads to the formation of immature virus particles that are transported to the Golgi apparatus.
The prM protein is cleaved in the trans-Golgi network (TGN) by host furin protease, completing the viral particle maturation. Finally, mature new virions are released from the host-cell by exocytosis.
BioNTech explains how dsRNA was increased
When the filthy Bacterial Process 2 was used without clinical trial to mass produce the Pfizer/BioNTech Covid19 Jab they told the EMA14 that they knew dsRNA would be increased as follows:
Therefore, all material additions into the system and their concentrations are known and chosen based on platform experience from BioNTech’s other RNA programs.
BioNTech transferred process 2 to Pfizer at lab scale, and process scale-up activities were then performed in parallel by BioNTech and Pfizer. Both companies aligned on the small scale and scale-up manufacturing processes.
For the current process, linearized plasmid DNA template was chosen for scalability in order to feasibly supply the current vaccine needs. The GTP and N1-methylpseudo UTP starting concentrations are controlled at a low target and these solutions are delivered as bolus feeds. These ribonucleotides were chosen to be limiting reagents to aid in capping and to reduce potential dsRNA impurities. The 5’-cap is in stoichiometric excess to the GTP to enable the preferential incorporation of the 5’-cap as the first addition to the RNA transcript. Additionally, controlling the N1-methylpseudo UTP concentration in the reaction is proposed to reduce the dsRNA impurities.
In parallel, small scale studies were conducted to understand the impact of a wider range of CTP and ATP, since ATP was identified as the next potential limiting nucleotide. Results showed that increasing the volumes of both CTP and ATP not only mitigated these nucleotide limitations, as measured at the end of the proteinase K step, but also increased integrity and yield and decreased dsRNA.
Process 1 and Process 2 DS batches show consistent removal of residual DNA template, dsRNA and other impurities. Residual DNA template in Process 1 batches was cleared to levels ≤ 200 ng/mg RNA and Process 2 batches have < 220 ng/mg RNA. Additionally, all clinical process 1 and process 2 batches were within specification acceptance criterion for dsRNA and comparably low (< 240 pg / microgram RNA).
New Zealand evaluators not satisfied
In response to the Pfizer EUA application NZ Health responded with a demand:
11) Provide a summary of the validation/verification status of the immunoblot analytical procedure used to detect double stranded RNA (dsRNA) in the active substance. Due date: March 2021.
Moderna Warned Shareholders about dsRNA
Profiteers from the mRNA jabs were warned by Moderna about the specific risks of their technology in compulsory filings with the US Securities Exchange Commission inncluding an expansive description on the last day of 2021.15
Our platform: mRNA science advancements
We continue to invest in both basic and applied research, seeking to advance both the state of our technology and the state of the scientific community’s understanding of mRNA. Examples of advances in mRNA science that combine nucleotide chemistry, sequence engineering, and targeting elements are described below.
mRNA chemistry: Modified nucleotides to mitigate immune system activation: The innate immune system has evolved to protect cells from foreign RNA, such as viral RNA, by inducing inflammation and suppressing mRNA translation once detected. Many cells surveil their environment through sensors called toll-like-receptors (TLRs). These include types that are activated by the presence of double-stranded RNA (TLR3) or uridine containing RNA fragments (TLR7, TLR8). Additionally, all cells have cytosolic double-stranded RNA, sensors, including retinoic acid inducible gene-I (RIG-I) that are sensitive to foreign RNA inside the cell.
The immune and cellular response to mRNA is complex, context specific, and often linked to the sensing of uridine. To minimize undesired immune responses to our potential mRNA medicines, our platform employs chemically-modified uridine nucleotides to minimize recognition by both immune cell sensors such as TLR3/7/8, and broadly-distributed cytosolic receptors such as RIG-I.
mRNA sequence engineering: Maximizing protein expression: mRNA exists transiently in the cytoplasm, during which time it can be translated into thousands of proteins before eventually being degraded. Our platform applies bioinformatic, biochemical, and biological screening capabilities, most of which have been invented internally that aim to optimize the amount of protein produced per mRNA. We have identified proprietary sequences for the 5’-UTR that have been observed to increase the likelihood that a ribosome bound to the 5’-end of the mRNA transcript will find the desired start codon and reliably initiate translation of the coding region. We additionally design the nucleotide sequence of the coding region to maximize its successful translation into protein.
Targeting elements: Enabling tissue-targeted translation: All nucleated cells in the body are capable of translating mRNA, resulting in pharmacologic activity in any cell in which mRNA is delivered and translated. To minimize or prevent potential off-target effects, our platform employs technologies that regulate mRNA translation in select cell types. Cells often contain short RNA sequences, called microRNAs or miRNAs, that bind to mRNA to regulate protein translation at the mRNA level. Different cell types have different concentrations of specific microRNAs, in effect giving cells a microRNA signature. microRNA binding directly to mRNA effectively silences or reduces mRNA translation and promotes mRNA degradation. We design microRNA binding sites into the 3’-UTR of our potential mRNA medicines so that if our mRNA is delivered to cells with such microRNAs, it will be minimally translated and rapidly degraded.
Impact of dsRNA on Epithelial Cells
In 2016 Moser and coworkers compared the effect of Endotoxin (LPS) with dsRNA on Human Umbilical Vein Endothelial Cells (HUVEC). Here is part of their Figure 2.
mRNA expression levels of genes in HUVEC treated with LPS (1 mg/ml) or transfected with RIG-I–specific agonist, 5’ 3p-dsRNA/LyoVec (2 μg/ml) for 4 and 24 h. Bars represent mean ± SD. All data are representative of three independent experiments. * ≤ 0.05, ** ≤ 0.01, *** ≤ 0.001.
Note that the reactions were time dependent and the significant increase in RIG-I and CXCL10.
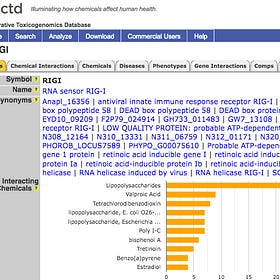
US CTD lists 1,794 Diseases linked to upregulated RIGI.16
These have had Human Curation. Click to magnify.
As mentioned before, US CTD lists 2,397 Diseases linked to upregulated CXCL10.17 These have had Human Curation. Click to magnify.
TGA reported dsRNA in Covid19 Jabs
Lot FL7649 “No More Than 40 picogram dsRNA/μg RNA” which is a little vague, perhaps reflecting their detection limit and not telling us if they busted open the LNPs to reveal the true content.
Somewhere I have seen more detail of TGA equipment and kit usd for dsRNA analysis of Covid19 Jabs, but will have to mine my database. Please feel free to help.
Queensland and NZ Jab Developers worry about dsRNA
Immunoblot analysis is judged inadequate for the extreme hazard posed by Picograms or less of dsRNA by Jab developer COVID19 Vaccine Corporation Limited (CVC) and collaborators in Queensland.18
If you have time, I recommend you read the reviewers comments and author responses.
Here is their immunoblot technique for dsRNA:
For the detection of dsRNA, the membrane was incubated overnight at 4 °C with two different dsRNA-specific murine monoclonal antibodies (mAb), which were derived from clones 3G1 and 2G4 (Mozzy Mabs, Brisbane, Australia). Both antibodies were incubated separately overnight at a 1:5 dilution, diluted in incubation buffer (1% Non-Fat Dry Milk in TBS-T). The membranes were then rinsed 3 times then washed 3 times for 15min each wash with TBS-T. The membranes were then incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG) secondary antibody (Abclonal, MA, USA) at 1:5000 dilution, diluted in incubation buffer, for 1 h with agitation. The membranes were then rinsed 3 times then washed 3 times for 15min each wash. Chemiluminescence detection was performed using Novex™ ECL chemiluminescent substrate kit (Invitrogen, MA, USA) and the signal intensities of the dots were visualised using the ChemiDocMP Imaging System (Bio-Rad, CA, USA).
Experts in the field might comment on use of Mouse IgG rather than Human IgG technology.
Here is their immunoblot analysis of dsRNA in the experimental NZ jab. You need a sharp eye to see they detected dsRNA using 2 different immunoblot antibody reagents.
dsRNA immunoblotting using two different dsRNA-specific antibodies to detect the presence of double-stranded RNAs in mRNA vaccine samples. The dsRNA positive control was generated from previous manufacture.
Recent paper on dsRNA and Pancreatic Cancer
The literature is exploding, so I just surveyed X to find some of the latest publications.
Here is the graphical abstract of one published on 21 November 2024 by Siyu Sun and coworkers19 showing how repeat units of dsRNA created by folding can overcome tumor suppression in Pancreatic Cancer.20
I will expand discussion when I have had time to read.
Further References Please
I know many Substack writers will have mentioned dsRNA, so please let me know of substantial discussion or any actual quantification in Covid19 or other Jabs.
Supyyyy. A computational model of Double-stranded RNA. Nucleic acid builder module of AMBERTools has been used to make this structure. Wikipedia.31 Julu 2020. https://en.wikipedia.org/wiki/RNA#/media/File:Double-stranded_RNA.gif
Poly(I:C) viral dsRNA mimic studies invalidated by Endotoxin Contamination
Readers are aware that I always look at the Materials and Methods section of papers to see where the reagents were sourced, and this led me to write about Endotoxin invalidating many Covid19 “Spikeopathy” studies of the Coronavirus and Jabs that encode synthetic Spike.
Leukaemia caused by Endotoxin in mRNA jabs
In recent articles I have shown that the Endotoxin contamination in all vials of the mRNA jabs causes a catalytic, positive feedback loop, increase in damaging microRNAs that can be directly linked to Heart and Liver Disease and Deaths, often in a very short time after the jab.
Legionnaires Bacteria Travel Long Distances in the Air in Victoria
Here is the Victorian Chief Health Officer, clearly worried about the latest Legionnaires Disease outbreak. It kills about one of every ten afflicted individuals.
53 years since Endotoxin and Lipid A were shown to be more toxic than dsRNA
In 1971 Endotoxin and its Lipid A were shown to be more toxic than dsRNA on a weight for weight basis, as shown in this figure.
https://dig-deeply.com/2024/03/15/foreign-dna-in-the-mrna-vaccine/
https://x.com/Kevin_McKernan/status/1858704704534180153
https://x.com/Patent_SUN
Septic Shock induced by Plasmid dsDNA and Truncated mRNA has common Sensing Mechanism to Endotoxin in mRNA Jabs
Many people have followed the excellent work of Dr Kevin McKernan analyzing samples of mRNA jabs foisted on billions of people worldwide to prove that the contamination includes fragments of the DNA grown inside E. coli bacteria from which the mRNA designed to code for a Stealth version of the Covid19 virus Spike Protein is made.
Érica Erlanny da Silva Rodrigues, Hannah Maus, Stefan Josef Hammerschmidt, Alessia Ruggieri, Elane Conceição dos Santos, Ênio José Bassi, Leticia Anderson, Pedro Gregório Vieira Aquino, João Xavier de Araújo-J únior, Fenju Wei, Xinyong Liu, Peng Zhan, Tanja Schirmeister, and Edeildo Ferreira da Silva-Júnior. The Medicinal Chemistry of Zika Virus. Human Viruses: Diseases, Treatments and Vaccines, https://doi.org/10.1007/978-3-030-71165-8_13
EMA 19 Dec 2019. Rapporteur Rolling Review critical assessment report. Quality aspects COVID-19 mRNA Vaccine BioNTech Modified mRNA, encoding full length SARS-CoV-2 spike protein. EMEA/H/C/005735/RR/xxx. Applicant: BioNTech Manufacturing GmbH
https://www.sec.gov/Archives/edgar/data/1682852/000168285222000012/mrna-20211231.htm
https://ctdbase.org/detail.go?type=gene&acc=23586&view=disease
https://ctdbase.org/detail.go?type=gene&acc=3627&view=disease
Helen M. Gunter, Senel Idrisoglu, Swati Singh, Dae Jong Han, EmilyAriens, Jonathan R. Peters, TedWong, SethW.Cheetham, JunXu, Subash Kumar Rai, Robert Feldman, Andy Herbert, Esteban Marcellin, Romain Tropee, Trent Munro and Tim R. Mercer. 2023. mRNA vaccine quality analysis using RNA sequencing. https://www.nature.com/articles/s41467-023-41354-y
Siyu Sun, Eunae You, Jungeui Hong, David Hoyos, Isabella S. Del Priore, Kaloyan M. Tsanov, Om Mattagajasingh, Andrea Di Gioacchino, Sajid A. Marhon,1 Jonathan Chacon-Barahona, Hao Li, Hua Jiang, Samira Hozeifi, Omar Rosas-Bringas, Katherine H. Xu, Yuhui Song, Evan R. Lang, Alexandra S. Rojas, Linda T. Nieman, Bidish K. Patel, Rajmohan Murali, Pharto Chanda, Ali Karacay, Nicolas Vabret, Daniel D. De Carvalho, Daniel Zenklusen, John LaCava, Scott W. Lowe, David T. Ting, Christine A. Iacobuzio-Donahue, Alexander Solovyov, and Benjamin D. Greenbaum. 2024. Cancer cells restrict immunogenicity of retrotransposon expression via distinct mechanisms. https://www.cell.com/immunity/fulltext/S1074-7613(24)00494-1
Pancreatic Cancer from Jabbing
I have mentioned various Jabs associated with Pancreatic Cancer, so it is time to collect what we know. It is the 4th leading cause of Cancer Death in the US.